install.packages('bigrquery')
library(bigrquery)
# authenticate
bq_auth()Google BigQuery and GCP (Google Cloud Platform)
Tutorial on how to set up BigQuery for your project using public datasets
Resource: Google BigQuery: First Steps in R
Create a GCP project
GCP organises resources by projects. If you do not already have a project, it’s time to create one!
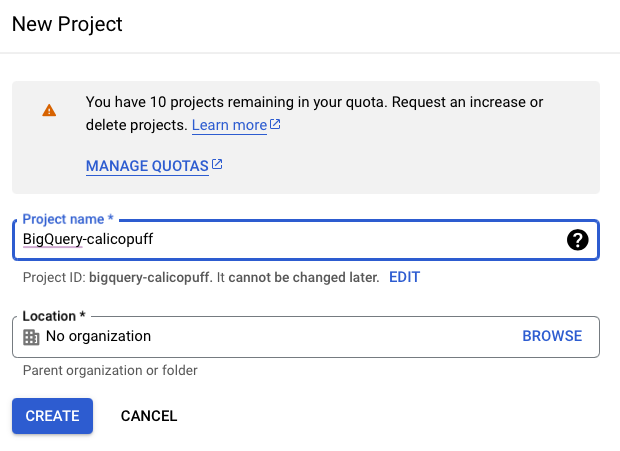
Create project with a name and ID
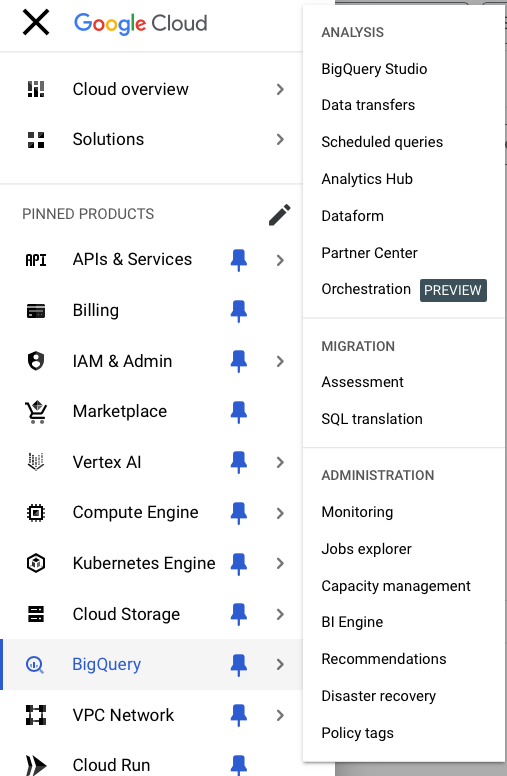
BigQuery in BigQuery Studio
Now go to BigQuery in the menu. Open BigQuery Studio.
Find Public datasets
Search for BigQuery resources
Can use the sources: public datasets

Now we use the examples of ADI (Area Deprivation Index) and Birth Data Summary schemas.
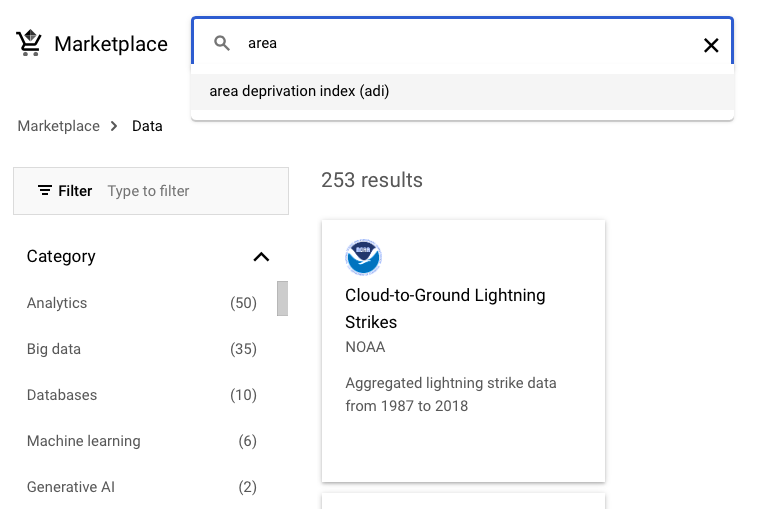
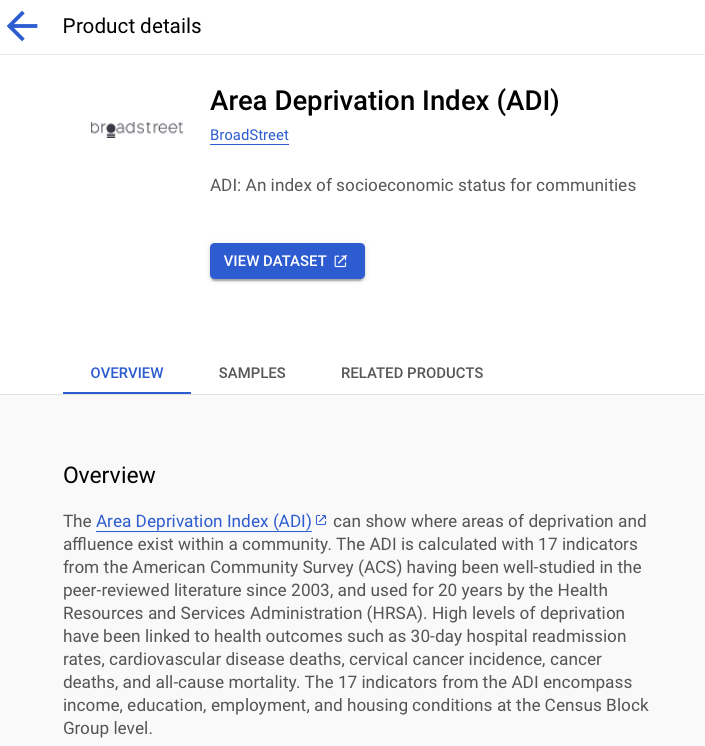
It is useful to star the dataset.
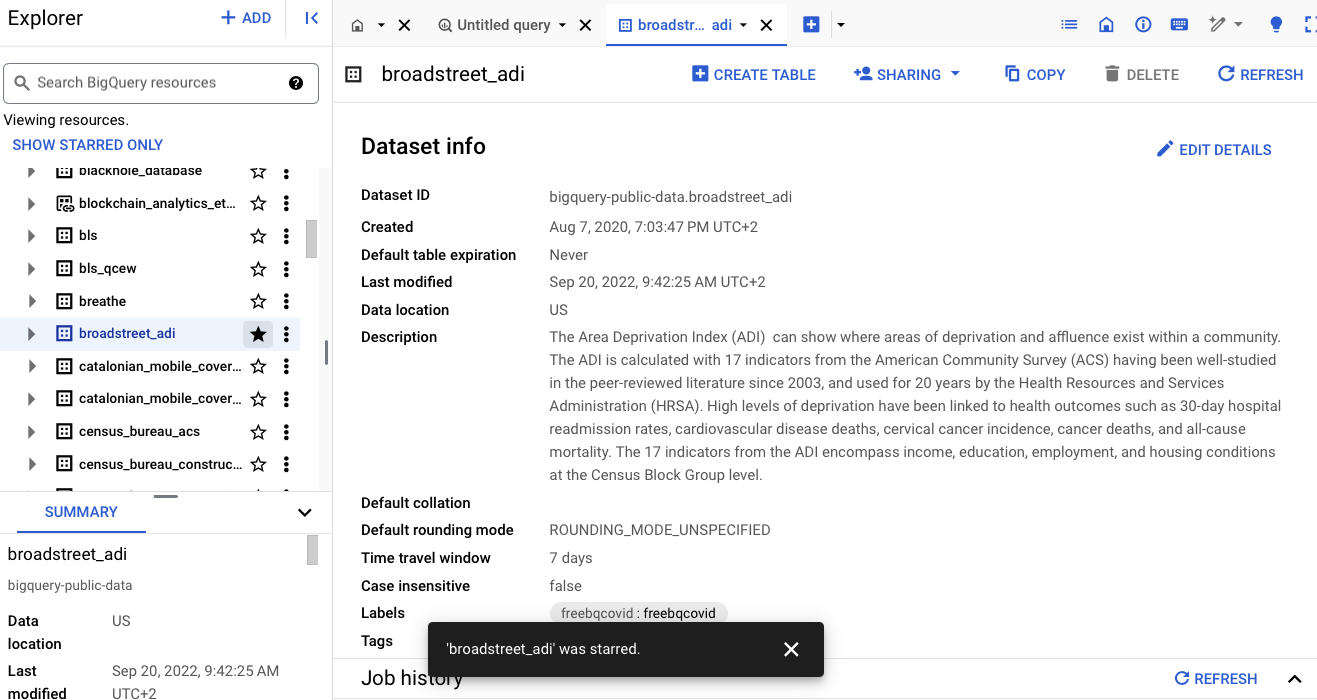
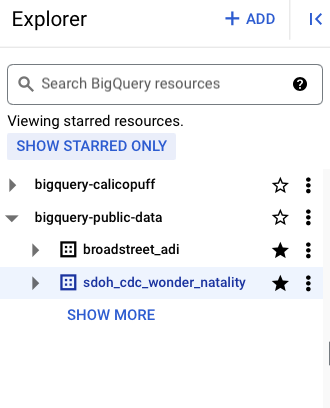
It is the entire schema (set of tables) that are added.
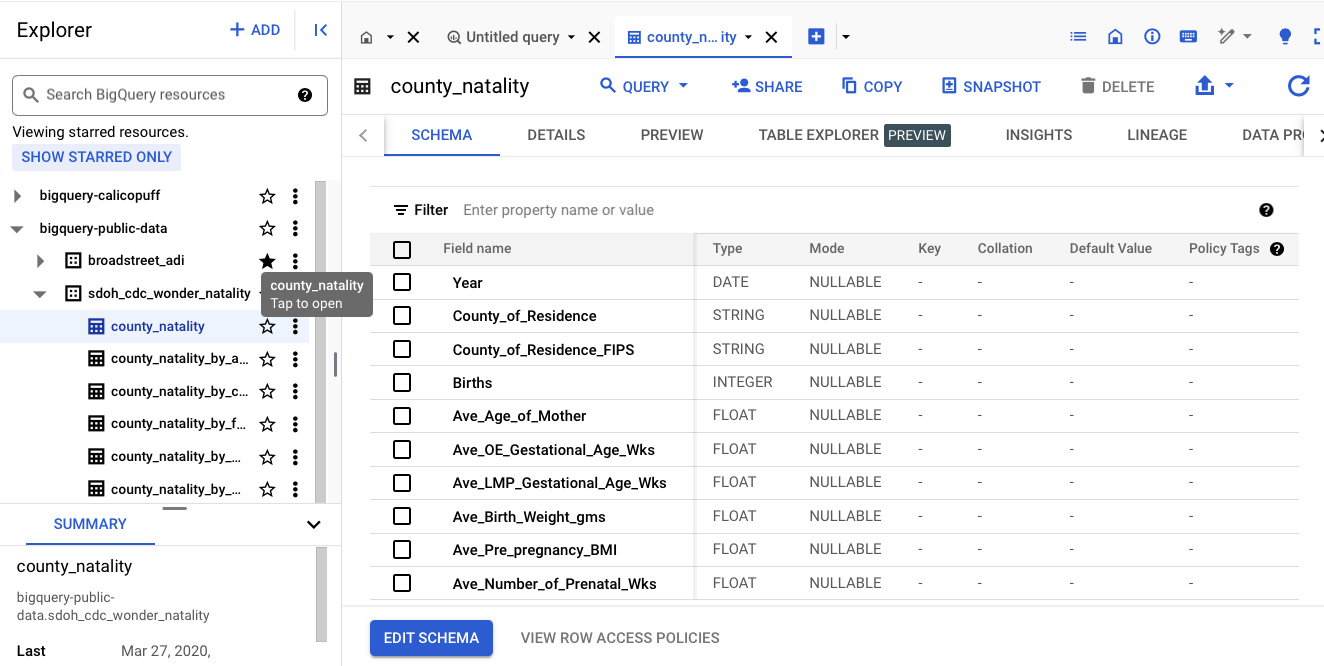
Can check the schema, details, preview to display the table.
Query
Either in a new tab, or a split tab.
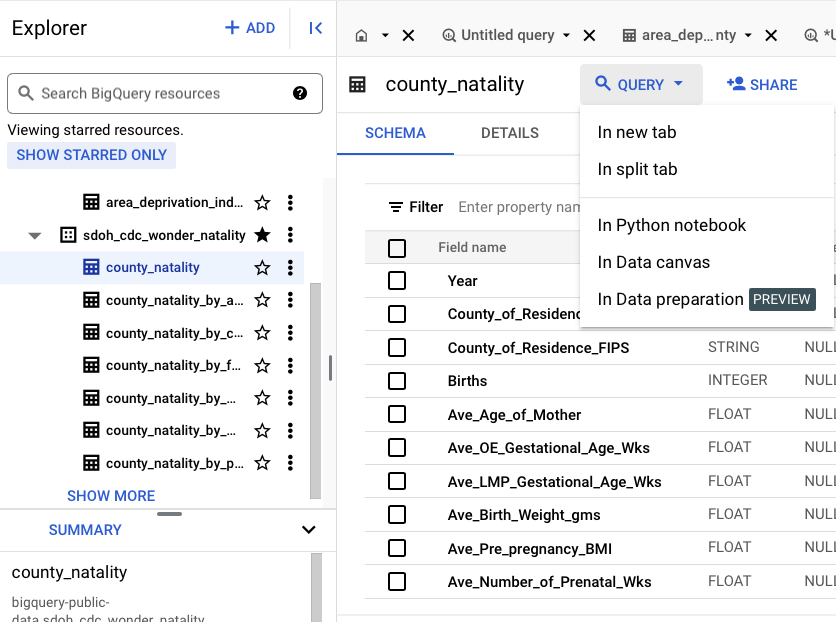
The query is pre-filled with some information. You can click on the schema column names to add it to the query.
Use fully qualified table names (project.dataset.table). It is good to limit the size of query results.
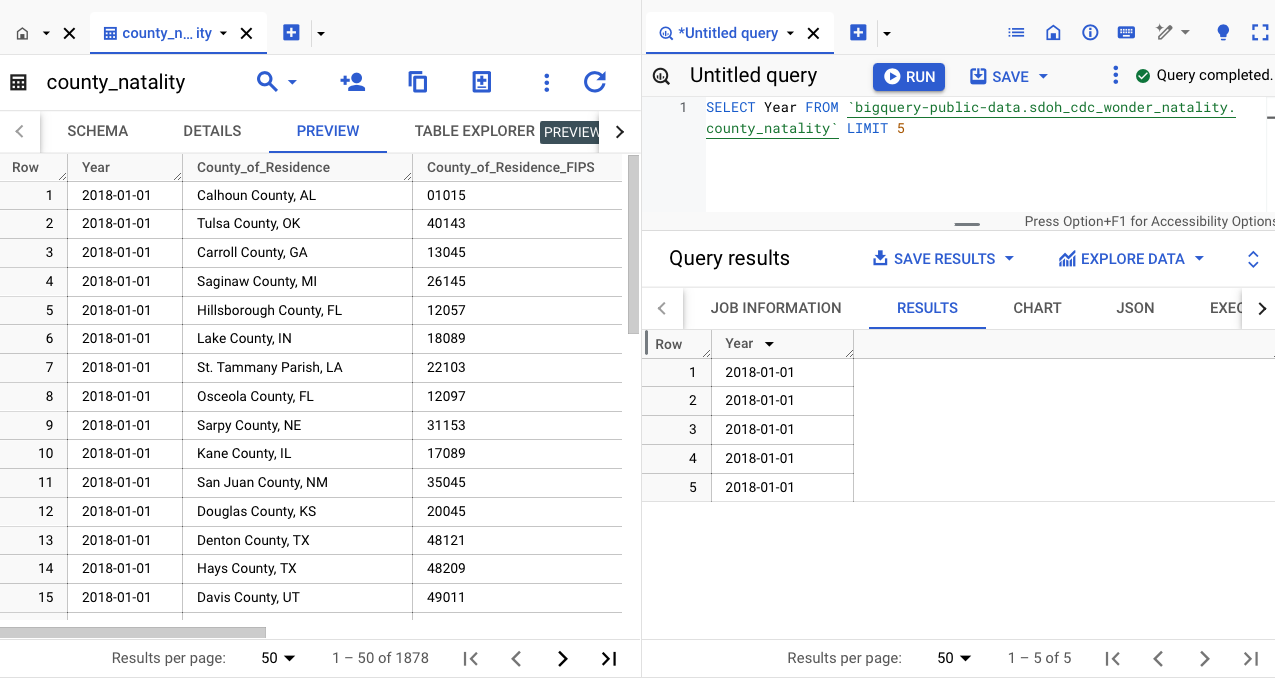
Join tables
When working on multiple tables, a good strategy to enable auto-filled fields is to select the tables first. On a second table (here for example it is county_natality), click on the menu (three dots), Copy ID - this will copy the fully qualified (long) table name.

It is good practice to use aliases.
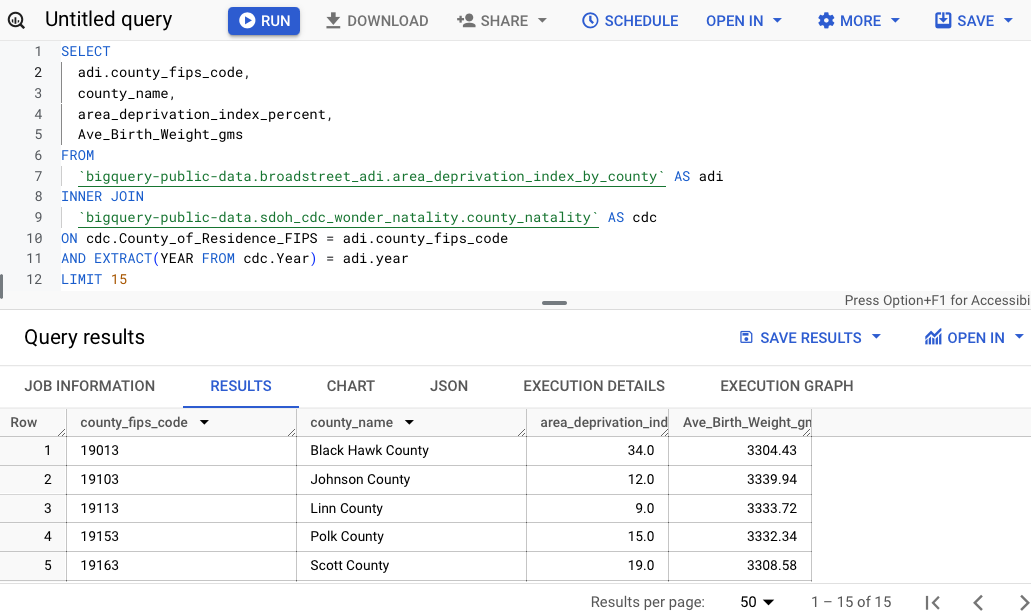
Save results
There are a few options to save the results:
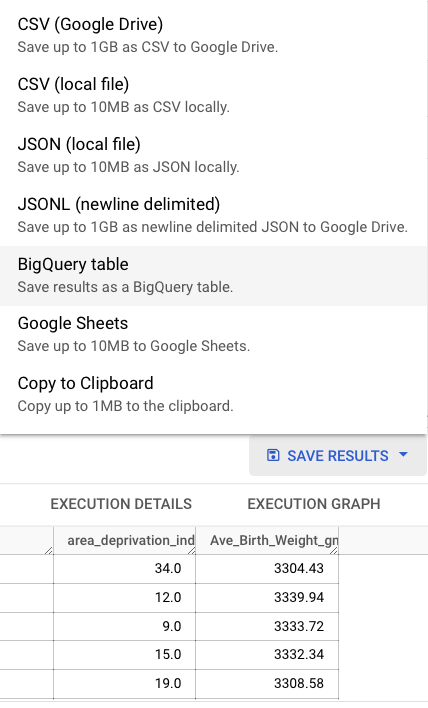
- save the query result (not table) locally: such as
csvfile - save BigQuery table on GCP
Note that the BigQuery table, once created, can not be downloaded as csv. You need to save the SQL query to reproduce it as query result in order to export it (or connect directly from R).
It is therefore important to save your SQL query!
Sometimes it is necessary to create a dataset in order to save the BQ table.
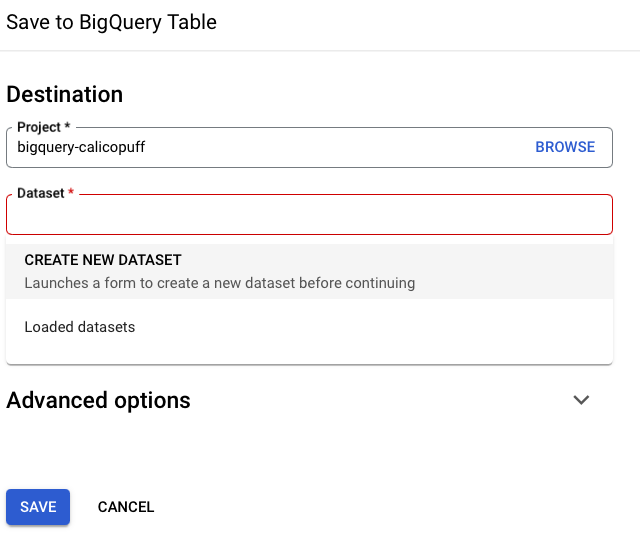
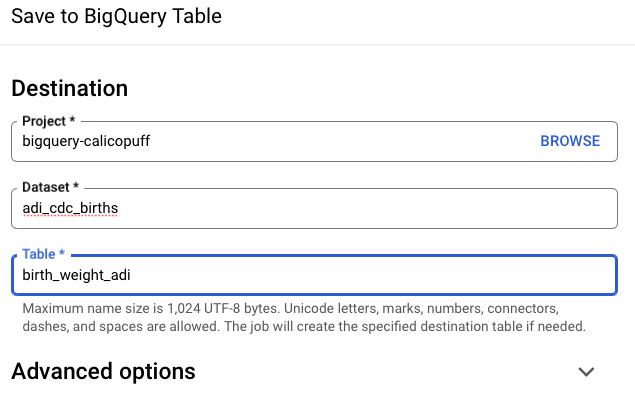
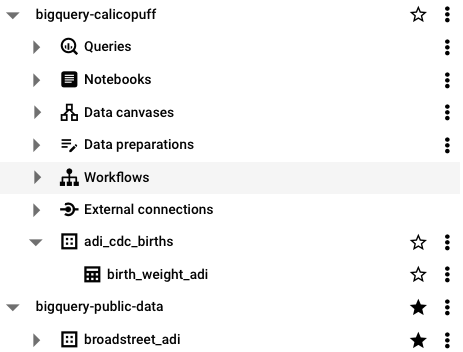
You can view the table in your own project list.
BigQuery in R
This section is an example using bigrquery package. Authentication is required.
Run a query from Rstudio”
- set
project_idandmy_sql_queryobjects - send query to BigQuery
results <- bq_project_query(project_id, my_sql_query) - get the table via
df <- bq_table_download(results)
project_id <- 'bigquery-calicopuff'
my_sql_query <- '
SELECT
date,
daily_confirmed_cases,
daily_deaths,
confirmed_cases
FROM `bigquery-public-data.covid19_ecdc.covid_19_geographic_distribution_worldwide`
LIMIT 300'Now that the project and query are defined, can run the code to extract the data.
results <- bq_project_query(project_id, my_sql_query)
df <- bq_table_download(results)